-Search query
-Search result
Showing 1 - 50 of 59 items for (author: frank & raw)
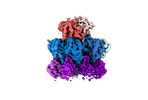
EMDB-19342: 
Human 20S proteasome assembly intermediate structure 5 beta7 tagged
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

EMDB-19343: 
Human 20S core particle beta7 tagged
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

EMDB-18755: 
Human 20S proteasome assembly structure 1
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA
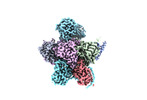
EMDB-18757: 
Human 20S proteasome assembly intermediate structure 2
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA
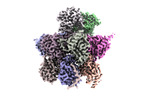
EMDB-18758: 
Human 20S proteasome assembly intermediate structure 3
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

EMDB-18759: 
Human 20S proteasome assembly intermediate structure 5
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA
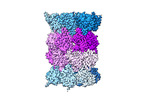
EMDB-18760: 
Human proteasome 20S core particle
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

EMDB-18761: 
Human preholo proteasome 20S core particle
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

EMDB-18762: 
Human premature proteasome 20S core particle
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

EMDB-18773: 
Human 20S proteasome assembly intermediate structure 4
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

PDB-8qyj: 
Human 20S proteasome assembly structure 1
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

PDB-8qyl: 
Human 20S proteasome assembly intermediate structure 2
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

PDB-8qym: 
Human 20S proteasome assembly intermediate structure 3
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

PDB-8qyn: 
Human 20S proteasome assembly intermediate structure 5
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

PDB-8qyo: 
Human proteasome 20S core particle
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

PDB-8qys: 
Human preholo proteasome 20S core particle
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

PDB-8qz9: 
Human 20S proteasome assembly intermediate structure 4
Method: single particle / : Schulman BA, Hanna JW, Harper JW, Adolf F, Du J, Rawson SD, Walsh Jr RM, Goodall EA

EMDB-18301: 
In-tissue cryo electron tomograms of App^NL-G-F amyloid plaques
Method: electron tomography / : Leistner C, Wilkinson M, Burgess A, Lovatt M, Goodbody S, Xu Y, Deuchars S, Radford SE, Ranson NA, Frank RAW

EMDB-16018: 
Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure
Method: helical / : Wilkinson M, Leistner C, Burgess A, Goodfellow S, Deuchars S, Ranson NA, Radford SE, Frank RAW

EMDB-16019: 
Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure (Methoxy-X04-labelled mice)
Method: helical / : Wilkinson M, Leistner C, Burgess A, Goodfellow S, Deuchars S, Ranson NA, Radford SE, Frank RAW

EMDB-4371: 
Cryo-EM map of the FCHSD2 F-BAR domain + SH3-1
Method: single particle / : Almeida-Souza L, Frank R, Garcia-Nafria J, Colussi A, Gunawardana N, Johnson CM, Yu M, Howard G, Andrews B, Vallis Y, McMahon HT

PDB-4v61: 
Homology model for the Spinach chloroplast 30S subunit fitted to 9.4A cryo-EM map of the 70S chlororibosome.
Method: single particle / : Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK

PDB-4v47: 
Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome
Method: single particle / : Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey ST, Sali A, Chapman MS, Frank J

PDB-4v48: 
Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome
Method: single particle / : Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey ST, Sali A, Chapman MS, Frank J

EMDB-1417: 
Cryo-EM study of the Spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins
Method: single particle / : Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK

EMDB-5116: 
30S subunit of ribosomal protein S1
Method: single particle / : Sengupta J, Agrawal RK, Frank J

PDB-2o0f: 
Docking of the modified RF3 X-ray structure into cryo-EM map of E.coli 70S ribosome bound with RF3
Method: single particle / : Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Liu Y, Zavialov A, Gursky R, Sanyal S, Ehrenberg M, Frank J, Song H

EMDB-1302: 
RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors.
Method: single particle / : Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Liu Y, Zavialov A, Gursky R, Sanyal S, Ehrenberg M, Frank J, Song H

EMDB-1362: 
Locking and unlocking of ribosomal motions.
Method: single particle / : Mikel V, Andrey Z, Sengupta J, Rawat U, Ehrenberg M, Frank J

EMDB-1363: 
Locking and unlocking of ribosomal motions.
Method: single particle / : Mikel V, Andrey Z, Sengupta J, Rawat U, Ehrenberg M, Frank J

EMDB-1364: 
Locking and unlocking of ribosomal motions.
Method: single particle / : Mikel V, Andrey Z, Sengupta J, Rawat U, Ehrenberg M, Frank J

EMDB-1365: 
Locking and unlocking of ribosomal motions.
Method: single particle / : Mikel V, Andrey Z, Sengupta J, Rawat U, Ehrenberg M, Frank J

EMDB-1366: 
Locking and unlocking of ribosomal motions.
Method: single particle / : Mikel V, Andrey Z, Sengupta J, Rawat U, Ehrenberg M, Frank J

EMDB-1184: 
Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM.
Method: single particle / : Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J

EMDB-1185: 
Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM.
Method: single particle / : Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J

PDB-2fvo: 
Docking of the modified RF1 X-ray structure into the Low Resolution Cryo-EM map of E.coli 70S Ribosome bound with RF1
Method: single particle / : Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J

PDB-2bcw: 
Coordinates of the N-terminal domain of ribosomal protein L11,C-terminal domain of ribosomal protein L7/L12 and a portion of the G' domain of elongation factor G, as fitted into cryo-em map of an Escherichia coli 70S*EF-G*GDP*fusidic acid complex
Method: single particle / : Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK

EMDB-1077: 
Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications.
Method: single particle / : Agrawal RK

PDB-1t1m: 
Binding position of ribosome recycling factor (RRF) on the E. coli 70S ribosome
Method: single particle / : Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J

PDB-1t1o: 
Components of the control 70S ribosome to provide reference for the RRF binding site
Method: single particle / : Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J

EMDB-1045: 
Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
Method: single particle / : Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J

EMDB-1006: 
A cryo-electron microscopic study of ribosome-bound termination factor RF2.
Method: single particle / : Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J

EMDB-1007: 
A cryo-electron microscopic study of ribosome-bound termination factor RF2.
Method: single particle / : Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J

EMDB-1008: 
A cryo-electron microscopic study of ribosome-bound termination factor RF2.
Method: single particle / : Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J

EMDB-1009: 
A cryo-electron microscopic study of ribosome-bound termination factor RF2.
Method: single particle / : Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J

EMDB-1010: 
A cryo-electron microscopic study of ribosome-bound termination factor RF2.
Method: single particle / : Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J

PDB-1pn6: 
Domain-wise fitting of the crystal structure of T.thermophilus EF-G into the low resolution map of the release complex.Puromycin.EFG.GDPNP of E.coli 70S ribosome.
Method: single particle / : Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J

PDB-1pn7: 
Coordinates of S12, L11 proteins and P-tRNA, from the 70S X-ray structure aligned to the 70S Cryo-EM map of E.coli ribosome
Method: single particle / : Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J

PDB-1pn8: 
Coordinates of S12, L11 proteins and E-site tRNA from 70S crystal structure separately fitted into the Cryo-EM map of E.coli 70S.EF-G.GDPNP complex. The atomic coordinates originally from the E-site tRNA were fitted in the position of the hybrid P/E-site tRNA.
Method: single particle / : Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J

PDB-1mvr: 
Decoding Center & Peptidyl transferase center from the X-ray structure of the Thermus thermophilus 70S ribosome, aligned to the low resolution Cryo-EM map of E.coli 70S Ribosome
Method: single particle / : Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
